3D Imaging of Cell’s Transport System May Shed Light on SMA, Other Disorders
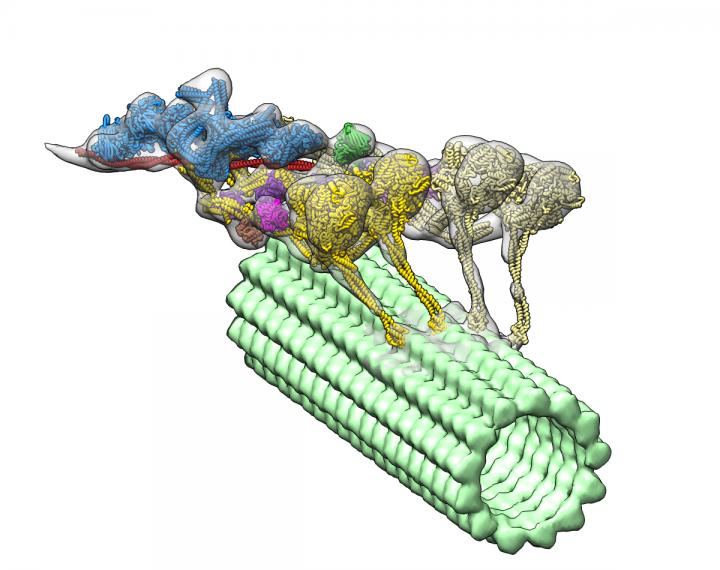
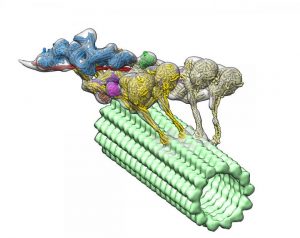
Using an advanced imaging technique and complex computational algorithms, researchers have created the first detailed, 3D imaging of a group of proteins called dyneins that may lead to a better understanding of the underlying causes of spinal muscular atrophy (SMA) and other neurodegenerative diseases.
Dyneins play a critical role in the cell’s transport system. Supported by structural elements called microtubules, they can travel throughout the cell delivering cellular cargo, such as signaling molecules and organelles (specialized structures within cells).
The study, led by researchers at The Scripps Research Institute (TSRI) and titled “Cryo-electron tomography reveals that dynactin recruits a team of dyneins for processive motility,” appeared in the journal Nature Structural & Molecular Biology.
Previous studies have reported that specific gene mutations affecting dyneins could lead to several disorders, including SMA and Charcot-Marie-Tooth (CMT) disease. However, it is not fully understood how these proteins contribute to the development of these diseases.
The new 3D image led researchers to the surprising discovery that another protein called dynactin actually hitches two dyneins together to form the protein complex, which enables them to deliver their large cellular cargo to the right destination.
“If you want a team of horses to move in one direction, you need to line them up,” Gabriel C. Lander, PhD, a TSRI associate professor and senior author of the study, said in a press release. “That’s exactly what dynactin is doing to dynein molecules.”
Researchers already knew that dyneins interacted with dynactins, but the discovery that two dyneins form the protein complex “was totally unexpected, and will change how this motor complex is represented in cell biology and biochemistry text books,” said Saikat Chowdhury, PhD, TSRI research associate and co-first author of the study.
To uncover the 3D structure of the dynein-dynactin complex, the team used an imaging technique called cryo-electron tomography. This is very similar to a CT scan, but applied to a protein. Then they developed advanced computational algorithms to interpret the collected information and reconstruct the flexible structure of the protein complex.
The discovery can help explain how dyneins transport heavy cargo, such as entire organelles, along microtubules over long distances in a crowded cellular environment. In addition, the imaging method the team developed can now be used to reveal the structure of other relevant protein complexes.
“We’re now able to move past cartoon models and visualize the fine details of many dynamic macromolecular complexes,” said Danielle Grotjahn, co-first author of the study. “As we learn more about the 3D organization and architecture of these molecular machines, we will be better equipped to understand how they malfunction in disease.”